Digital PCR (dPCR): Ultra-Sensitive Detection of MAP
Introduction
Digital PCR (dPCR) is one of the most advanced molecular techniques used today for detecting and quantifying DNA. Unlike traditional PCR, which provides relative quantification, dPCR offers absolute and highly sensitive measurement of Mycobacterium avium subspecies paratuberculosis (MAP) DNA even at very low concentrations. This makes it a powerful tool for early diagnosis and monitoring of paratuberculosis in livestock.
How Does Digital PCR Work? 🔬
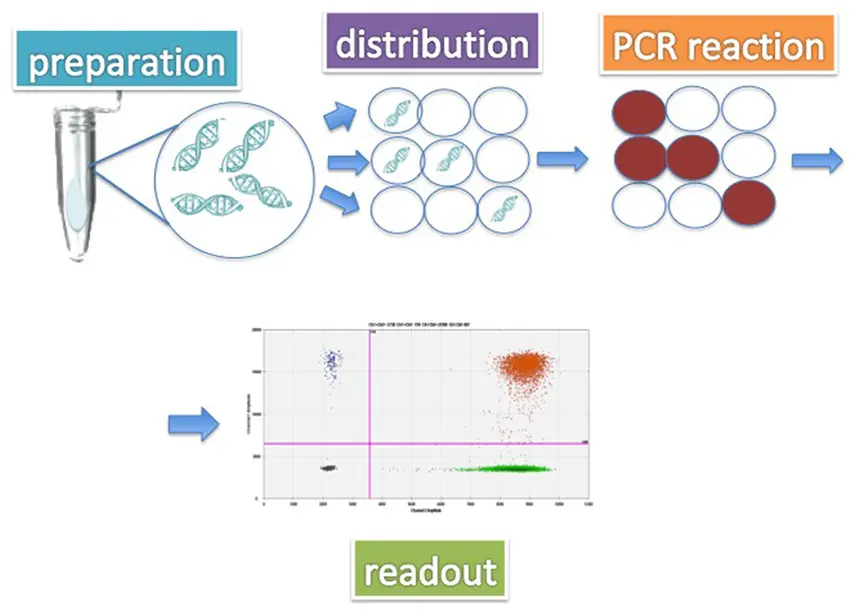
Digital PCR partitions a DNA sample into thousands to millions of tiny, separate reactions. Each partition ideally contains either zero or one target DNA molecule. PCR amplification then occurs independently in each partition. By counting how many partitions yield positive results, dPCR calculates the exact number of DNA copies in the original sample.
- Partitioning: The sample is divided into thousands of tiny droplets or wells.
- Amplification: PCR runs independently in each droplet/well.
- Detection: Positive partitions emit a fluorescent signal, which is counted.
- Quantification: The ratio of positive to total partitions provides an absolute DNA count.
Advantages of dPCR for MAP Detection 🎯
- Extreme Sensitivity: Can detect very low levels of MAP DNA, critical for early infection stages when bacteria counts are low.
- Absolute Quantification: No need for standard curves or references; provides direct copy number measurement.
- Resistance to Inhibitors: More tolerant to substances that may inhibit PCR, improving accuracy in complex samples like feces or soil.
- Reproducibility: Offers consistent results even at low bacterial loads.
- Precise Monitoring: Useful for tracking bacterial load changes over time in infected animals or environmental samples.
Applications in Paratuberculosis Research and Control 🐄
- Early Diagnosis: Detects MAP infections before clinical symptoms appear, allowing earlier intervention to limit spread.
- Herd Health Monitoring: Enables quantitative tracking of infection levels within livestock populations.
- Environmental Surveillance: Monitors MAP presence in soil and water around farms to better understand contamination and persistence.
- Research: Supports studies on MAP transmission dynamics, antibiotic resistance, and bacterial persistence.
Technical Aspects and Workflow ⚙️
- Sample Preparation: Extraction of DNA from blood, feces, milk, or tissue samples.
- Partitioning: Using droplet generators or microfluidic chips to separate the DNA sample.
- PCR Amplification: Running thermocycling steps with target-specific primers and probes.
- Detection: Fluorescence detection instruments count positive partitions.
- Data Analysis: Software calculates absolute DNA concentration and provides detailed reports.
The Future of dPCR in Paratuberculosis Control 🔮
With ongoing technological improvements, dPCR is becoming more accessible and faster, enabling field-friendly platforms and integration with digital data management systems. The ability to quantify MAP DNA precisely will continue to transform disease surveillance, control programs, and scientific research bringing us closer to effective management of paratuberculosis worldwide.