📌 Introduction
Illumina sequencing, commonly referred to as Next-Generation Sequencing (NGS), is a popular platform that provides cost-effective outcomes, high accuracy, and massive throughput. It is essential to comparative genomics, diagnostics, and population studies in MAP (Mycobacterium avium subsp. paratuberculosis) research.
🔧 How It Works
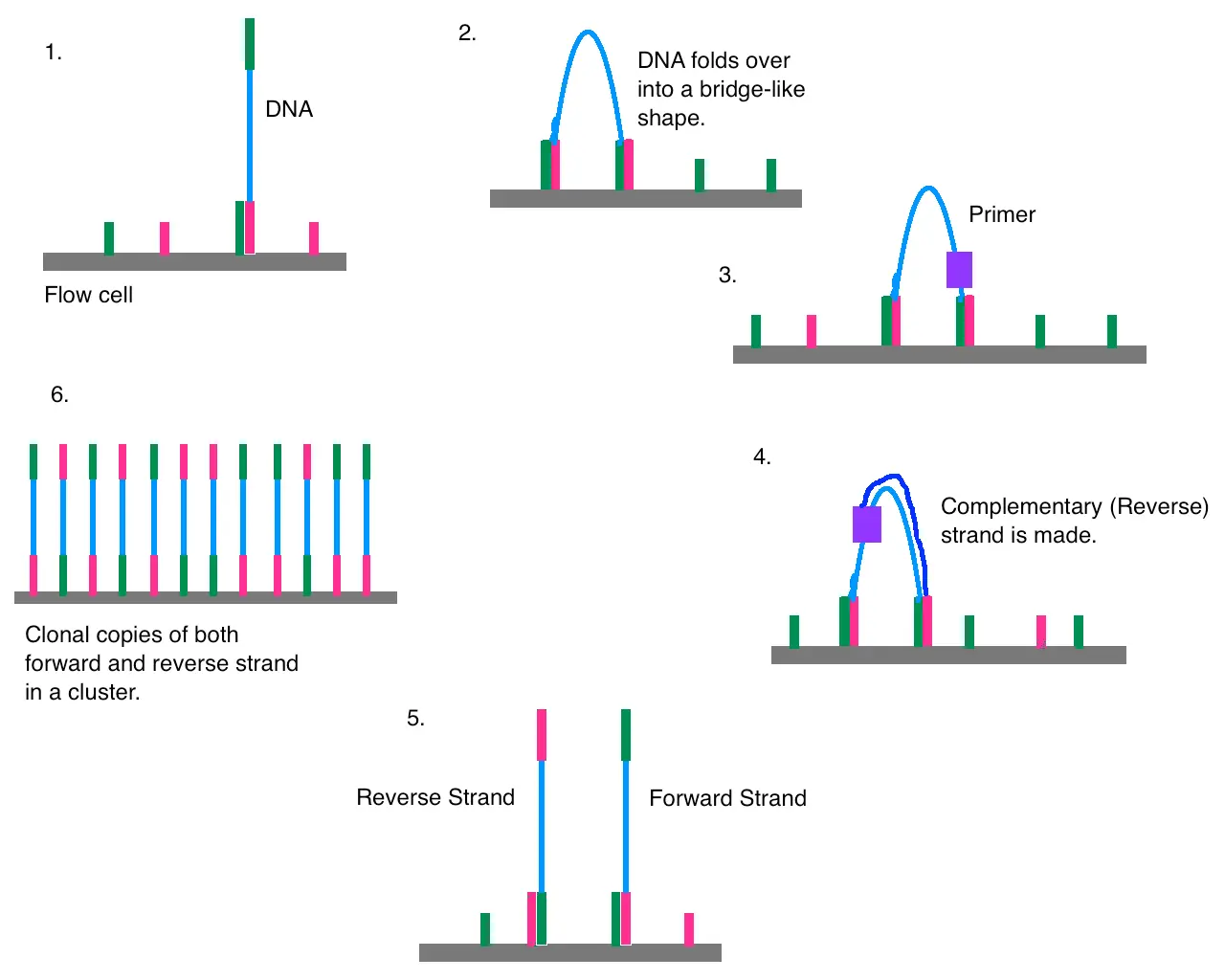
Illumina sequencing uses a method called sequencing-by-synthesis:
- DNA is fragmented and ligated to adapters.
- Clusters are formed on a flow cell.
- DNA polymerase incorporates fluorescently labeled nucleotides, which are recorded in real-time.
It generates short, high-quality reads (typically 50–300 bp) that can be assembled or aligned to reference genomes.
🎯 Key Benefits for MAP Studies
| Feature | Benefit for MAP Research |
|---|---|
| 🎯 High Accuracy | Reduces sequencing errors, especially critical in identifying SNPs and resistance genes. |
| 📊 Massive Throughput | Allows sequencing of hundreds of MAP samples simultaneously—ideal for large epidemiological studies. |
| 💲 Cost Efficiency | Lower cost per base compared to many other platforms, especially when processing many samples. |
| 🔍 SNP Detection | Detects single-nucleotide polymorphisms crucial for tracking MAP evolution or drug resistance. |
🧬 Applications in MAP Research
🧩 Whole-Genome Resequencing (WGR)
Perfect for mapping known MAP strains and identifying mutations, rearrangements, or deletions across populations.
🧬 Metagenomics
Used to identify MAP directly in complex microbiome samples like feces, milk, or soil—without prior culturing.
📌 Targeted Amplicon Sequencing
Focus on specific genes (e.g., IS900, f57, hspX) to quickly confirm MAP presence and identify strain types.
🧬 Transcriptomics (RNA-seq)
Reveals how MAP expresses its genes in response to stress, host immunity, or during infection—critical for understanding virulence mechanisms.
🧪 Illumina-Compatible Tools for MAP Research
| Tool | Purpose |
|---|---|
| BWA | Aligning reads to reference genome |
| GATK | Variant calling and SNP analysis |
| SPAdes | Genome assembly |
| MetaPhlAn | Metagenomic profiling |
| Salmon / Kallisto | Quantifying gene expression (RNA-seq) |
🧠 Why Add Illumina to Your MAP Toolbox?
Illumina excels when accuracy and scale are critical. Whether you're studying hundreds of MAP genomes, analyzing gene expression, or conducting diagnostic surveillance, this platform is a gold standard in molecular microbiology.
It complements other technologies like:
- Oxford Nanopore (for long-read scaffolding),
- qPCR or dPCR (for routine diagnostics),
- Proteomics (to validate gene expression at the protein level).